FAQs and common problems
Installation
Do I need to edit the package setup script?
You may well have to edit the setup script depending on the target system. This includes editing compiler flags (see below for example regarding instruction sets).
Does it matter what compiler I use?
The Intel compiler collection has been used successfully for X-PSI and dependencies (namely GSL, MultiNest). We recommend first trying to use Intel in a context where performance matters.
What Intel instruction sets should I use?
If you want to test the binaries on a login node, note that you can
compile with multiple instruction sets for auto-dispatch using the -x and
-ax flags. See the HPC systems page for examples.
Sampling
Is I/O or disk storage a concern, or are all the files small?
I/O not a concern for likelihood calculation.
Nested sampling writes to disk at user-specified cadence (so many nested sampling iterations).
Model data such as a four-dimensional atmosphere table can be reasonably large for I/O. We recommend loading, at the outset of the run (or a resumed run), such a table into a contiguous chunk of memory for each of the Python processes running on one node. That table is pointed to for access where needed from compiled modules (C extensions to Python): it is not loaded from disk per likelihood call. We provide an example custom Python class that handles this loading (as used in Riley et al. 2019, hereafter R19).
Disk storage required is indeed small: up to \(\mathcal{O}(100)\) Mbytes for applications thus far (e.g., R19). There is a variant of MultiNest nested sampling that is much more memory and disk intensive, but we do not use it. This is because importance nested sampling is not compatible with the alternative options (read: hacks) for prior implementation (see Riley, PhD thesis).
Common problems and errors
How to avoid errors in post-processing?
Do not use X-PSI PostProcessing tools for runs which have not converged yet or have not enough samples. Also, when post-processing, make sure to check the data and output file paths, use cache=True if plotting the signal, and perform a likelihood check to be sure that the imported model is the same as in the run.
AttributeError: ’NestedBackend’ object has no attribute ’:math:`_nc_bcknd`’
Can happen in PostProcessing for runs with use_nestcheck=[False] (e.g. importance sampling). Solution is to turn bootstrap_estimators=False, or alternatively, set use_nestcheck=[True].
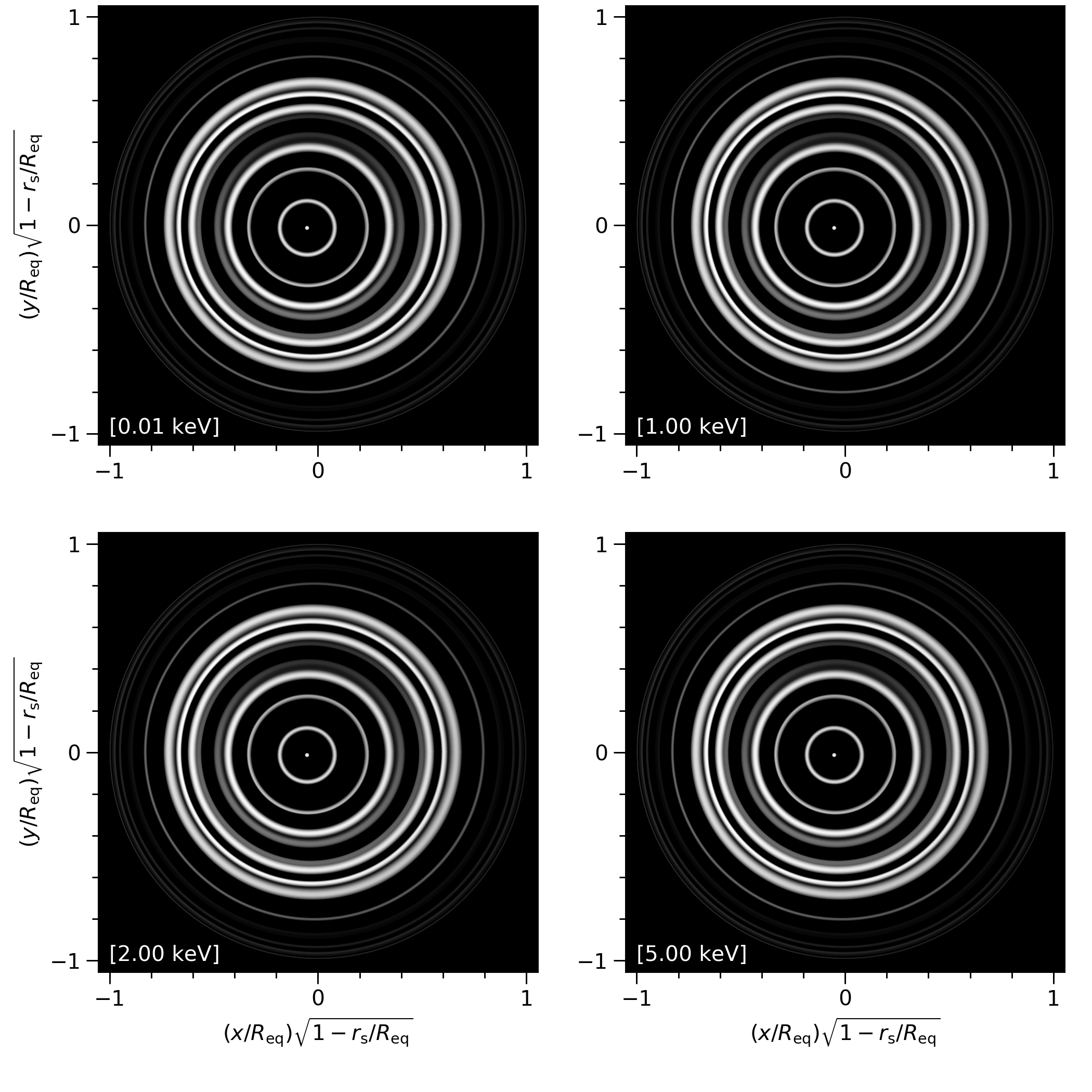
Why does my skymap show many annular images like this:
The problem is the xpsi/xpsi/surface_radiation_field/local_variables.pyx file which should be overwritten by xpsi/xpsi/surface_radiation_field/archive/local_variables/PST_U.pyx or xpsi/xpsi/surface_radiation_field/archive/local_variables/two_spots.pyx (depending on the model) and then re-install X-PSI.
ImportError: No module named tools
You are running X-PSI from its main directory (the directory where the setup.py file is). Exit that directory and run it again.
<path/to/run/output>dead-birth.txt not found.
Set use_nestcheck=[False] or check that nestcheck is installed exactly as instructed in Installation (by cloning it from https://github.com/ThomasEdwardRiley/nestcheck.git).
Invalid caching targets.
Set cache=True for the signal.
Each row and column must contain at least one positive number.
There are some rows and/or column in the instrument response that contain only zeros. Solution is to increase the number of channels or decrease the number of energy intervals.
Warning: Using native nestcheck KDE instead of GetDist KDE.
Make sure to to install nestcheck and GetDist packages using the corresponding github repositories as instructed in Installation.
ValueError: There is more than one signal instance.
Typically occurs when post-processing joint NICER and XMM results, if not setting model.likelihood.signals = model.likelihood.signals[0][0] (when plotting the inferred NICER signal).
Weird issues
The import order of pymultinest and numpy may sometimes affect the seed-fixed results.
The import order of numpy and X-PSI (and thus pymultinest) was observed to sometimes influence the exact sampling results even if fixing the random seeds for both numpy and pymultinest. However, the issue occurred only in specific conda environments (with certain packages/dependencies) and later attempts to reproduce it with newer X-PSI installations have not been successful. See the GitHub issue for more details.
Linker library name format in setup.py may cause issues with install under rare circumstances.
setup.py has the following if-else block (twice):
if 'clang' in os.environ['CC']:
libraries += ['inversion.so', 'deflection.so']
else:
libraries += [':inversion.so', ':deflection.so']
The : indicates to the GNU linker that the library name should be taken as is, that is, not prepended with lib. [1]
The linker on Darwin (macOS), probably a BSD(-derived) linker, takes any(?) name that has a extension as the literal name, and won’t search for lib<name>.dylib or lib<name>a. [2] So that linker does this automagically, whereas the GNU linker wants to be explicit.
Note the use of linker in the above paragraph: it is not the compiler that makes the difference, but the linker. On the command line, it will look like the compiler (the linking step still shows the compiler being used), but under the hood it calls the linker, ld. Therefore, the linker should be the differentiating factor which of the two if-branches is used, not the compiler (or its environment variable, CC).
This issue pops when building X-PSI on macOS with gcc (note: not the built-in gcc, which is clang in disguise: check gcc --version; but for example gcc installed with Homebrew), or when using clang on Linux. Both builds will error at this point when linking with the so-files is done.
A manual, local, quick fix is just to insert or remove the colons in front of the library names in setup.py, and the build can continue. But this is only a local solution, and can’t be implemented in setup.py by default.
The linker, however, is trickier to determine than the compiler. But the linker is also tied into the system ecosystem (through system libraries); for example, the GNU linker is not supported on Darwin (though perhaps a BSD linker can be installed on a Linux system).
Instead, one could try and determine the system OS and use that, instead of the compiler, in the if-statement: sys.platform may be of use here.
Note that the use cases will be rare: the default compiler on macOS tends to be clang (installed as part of the XCode tools), and even the default gcc is just clang in disguise. On Linux, the majority of times the default compiler is gcc. I don’t know which linker the Intel compiler will use; perhaps the GNU linker, perhaps it has its own linker (but given that the OS is formally GNU/Linux, I suspect the linker is the GNU one). Most people are not likely to deviate from this, so the check for clang tends to nearly always be True for macOS and False for Linux.
Footnotes